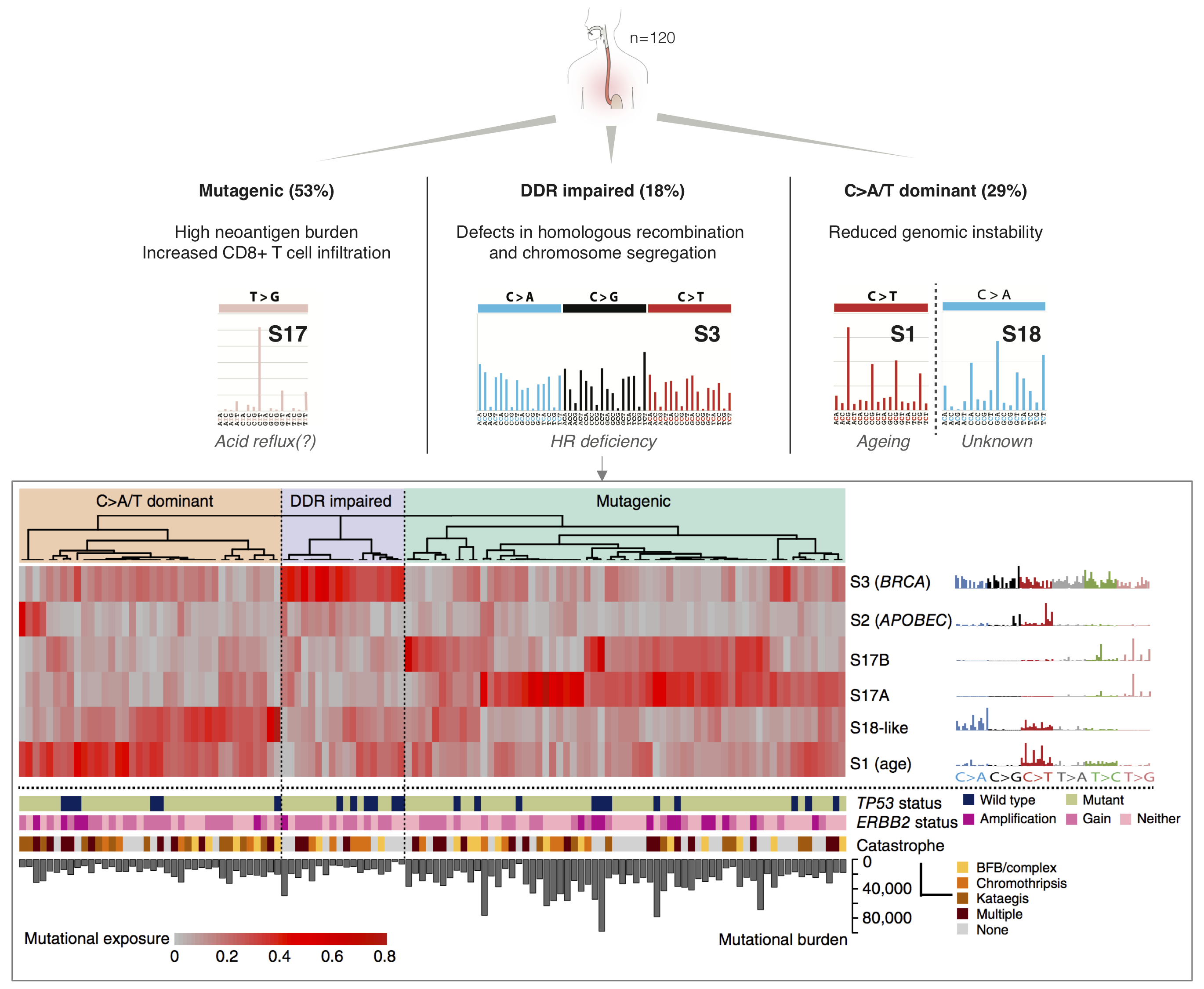
We have previously shown that mutational footprints associated with extrinsic and intrinsic risk factors of cancer can be used to inform aetiology and treatment options in oesophageal adenocarcinoma (Secrier, Li et al, Nat Genet 2016). Specifically, we uncovered three distinct subtypes of this cancer with distinct therapeutic opportunities, including a subgroup with high mutational burden, which could benefit from immunotherapy, and one with prevalent DNA damage defects in the homologous recombination pathway, for which PARP inhibitors may be effective. Moreover, we have characterised the dynamics of mutational processes across disease stages, from Barrett Oesophagus to primary tumours and metastases (Abbas et al, Nat Commun 2023).
We are currently investigating further mechanisms of mutational signature development and their involvement in cancer progression.
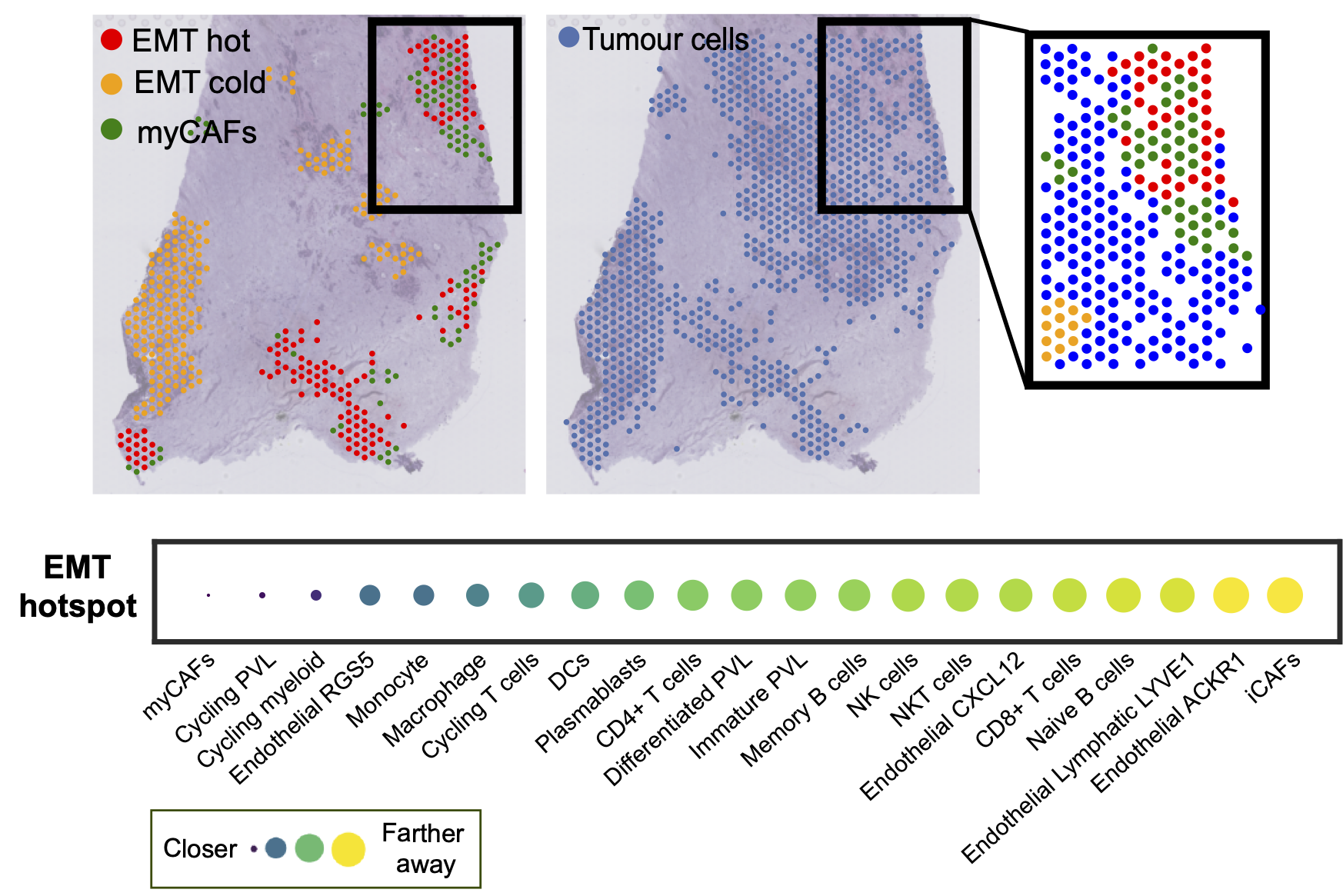
We are interested in developing new methodology for evaluating cancer-immune cell interactions and immune activity as determined by cancer cell plasticity.
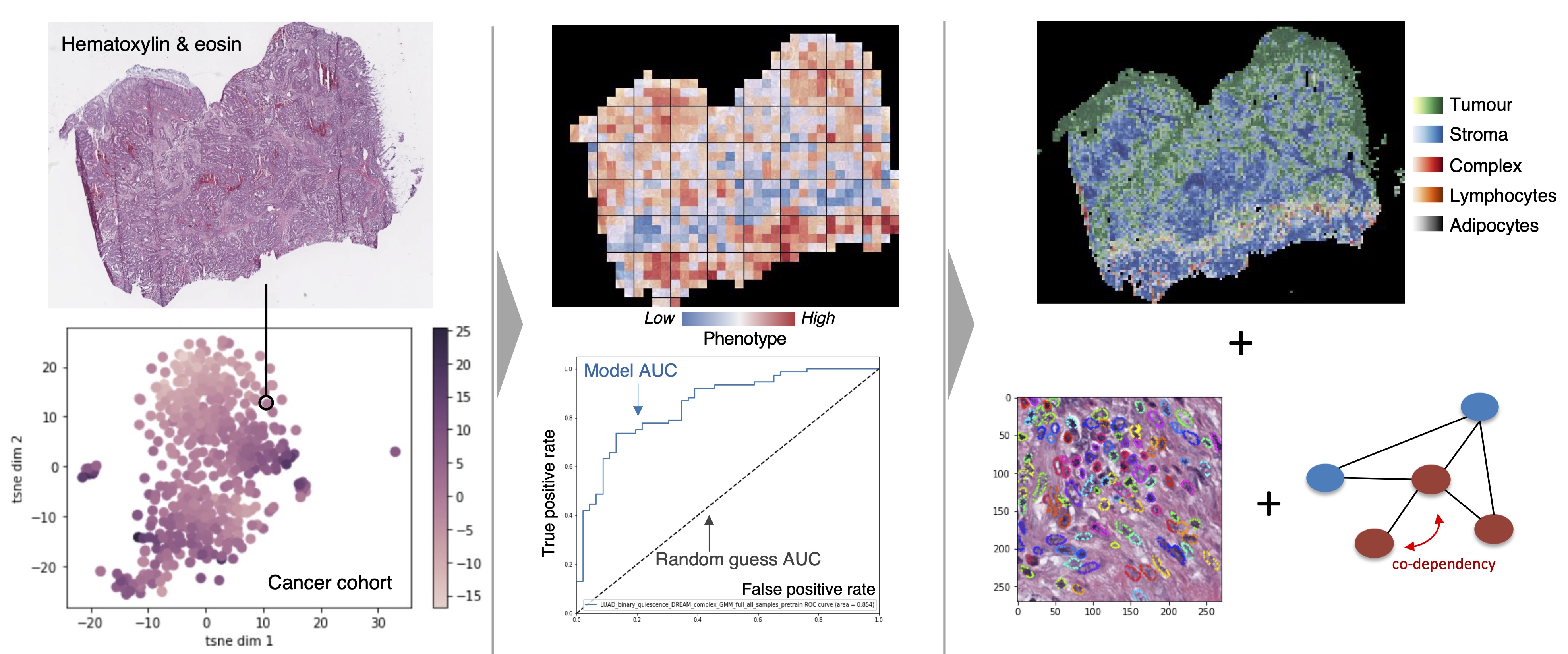
We employ deep learning and graph-based methodology to discover cell states and phenotypes from cancer histopathology images. We are developing multi-omics and imaging data integration approaches that enable us to explore tumour dormancy, the epithelial-to-mesenchymal transition and the role of the tumour microenvironment in cancer progression.
We apply unsupervised and supervised learning approaches for the purpose of patient stratification and prediction of disease outcome or response to therapy. At the cellular level, we apply similar methods to understand dependencies within protein interaction networks and the effect of signal propagation within key pathways regulating oncogenic activation, senescence, dormancy, metastasis etc.
We are developing new tools for multi-omics data integration. Methods employed include: causal reasoning, predictive models, network analysis, hotspot/module identification etc.